Different types of RNA,s and their functions
In this article we discussed about different types of rna and their functions in detail. like tRNA, mRNA, rRNA, hnRNA, siRNA etc

Table of Contents
Introduction to RNA
While DNA is the hereditary material of eukaryotes which contain all the information needed to maintain a cell’s processes, but these do not leave the nucleus. So, transmission of data from the nucleus to body of the cell is mediated by RNA, the ribonucleic acid. RNA molecules play essential
role in transfer of genetic information during protein synthesis and in regulation of gene expression. For some viruses and viroids, RNA act as storage of genetic information. Hence, RNA molecules may be genetic or non genetic, catalytic (like ribozymes) or non catalytic and coding (like mRNA) or
noncoding (like tRNA and rRNA). In 1967 Carl Woese found the catalytic properties of RNA and speculated that the earliest forms of life relied on RNA both to carry genetic information and to catalyze biochemical reactions

What are the chemical elements of RNA?
<div jsname="Q8Kwad" style="background-image: url("data:image/svg+xml,"); display: inline-block; height: 24px; width: 24px; color: rgb(32, 33, 36); font-family: arial, sans-serif; font-size: medium; font-weight: 400; white-space-collapse: collapse;">
- RNA is usually a single-stranded molecule, while DNA is double stranded.
- It consists of four nitrogenous bases: adenine, cytosine, uracil, and guanine. Uracil is a pyrimidine that is structurally similar to the thymine, another pyrimidine that is found in DNA. Like thymine, uracil can base-pair with adenine.
- Also, the sugar in RNA is ribose instead of deoxyribose (ribose contains one more hydroxyl group on the second carbon), which accounts for the molecule’s name
- Since RNA is single stranded complementary to one of the strand of DNA, neither its guanine content is necessarily equal to cytosine nor the adenine content equals the uracil content.
- Alkali lability of RNA yielding 2’-3’ cyclic diesters of mononucleotides is useful both diagnostically and analytically,which is not possible in case of DNA due to absence of 2’ hydroxyl group.
What are different types of RNA
Within a given cell, RNA molecules are found in multiple copies and multiple forms. Major RNA classes are mRNA, tRNA and rRNA
Messenger RNA (mRNA)
- carries the genetic information copied from DNA in the form of series of triple codons (three base code words), each of which specifies specific amino acid.
- Hence, it act as a template for protein synthesis.
- mRNA can range from ~300 nucleotides to ~7000 nucleotides, depending on the size and the number of proteins that they are coding for.
- Most of the eukaryotic mRNAs represent only a single gene as they are monocistronic.
- Most of the prokaryotic mRNA are polycistronic as they carry sequences coding for several proteins. In such cases, a single mRNA is transcribed from a group of adjacent genes.
- All mRNAs contain two types of regions. The coding region consist of a series of codons starting with an AUG and ending with termination codon UAG, UAA, and UGA.
- Apart from this, extra regions are present at both ends. The untranslated region at 5’end is called leader and untranslated region at the 3’ end is called trailer.
- Eukaryotic mRNA molecules often require extensive processing and transport the Processing involves capping (methylation at 5’ end), introns (non coding regions) removal and tailing (polyadenylation at 3’end).
Ribosomal RNA (rRNA)
- Ribosomal RNA (rRNA) which make up an integral part of the ribosome.
- The ribosome is large protein synthesis machinery in the cell (~ 20 nm in diameter, 70S sedimentation rate for bacterial ribosomes and 80s for eukaryotes).
- Bacterial ribosomes are made up of two subunits: a large subunit (~50S) and a small subunit (~ 30S).
- The large subunit is in turn made of two ribosomal RNA (5S and 23S) and several (~34 proteins) whereas the small subunit has one ribosomal RNA (16S) and ~ 21 proteins.
- The 23S rRNA is ~ 3000 nucleotides long, and the 16S rRNA is ~ 1500 nucleotides long.
- On the other hand eukaryotic ribosome like that of mammals contain two major nucleoprotein subunits-larger one (~60S) and small one (~40S).
- The large subunit is in turn made of three ribosomal RNA (5.8S, 5S and 28S r RNA) and more than 50 proteins) whereas the small subunit has single ribosomal RNA (18S) and ~30 proteins.
- All of the rRNAs except 5S r RNA are processed from a single 45s precursor RNA molecule in the nucleolus.
- Highly methylated rRNA molecules are packaged in the nucleolus with specific ribosomal proteins.
- The function of rRNA is quite unclear but they are necessary for ribosome assembly and seem to play important role in binding of mRNA to ribosomes and its translation.
- According to some recent researches, rRNA component performs the peptidyl transferase activity and thus have intrinsic catalytic nature involving cleavage of nucleic acid.
- There are large number of ways in which hairpins and loops can be formed in rRNA. Its structure prediction requires a combination of both computational methods (in which the most probable secondary structures are determined from estimates of free energy for a given structure) and a variety of experimental techniques.
Transfer RNA (tRNA)
- Transfer RNA (tRNA), is the adapter molecule mainly responsible for decoding mRNA into the corresponding peptide sequence.
- The many forms of tRNA have roughly the same size and shape, varying from about 73 to 93 nucleotides.
- Besides the usual bases A, U, G, and C all have a significant number of modified bases, which are thought to have been formed by modification after the transcription.
- Mahlon Hoagland in 1956 discovered transfer RNA. The sequence of the 77 nucleotides of yeast tRNA was found by Robert W. Holley in 1965 and Holley won the 1968 Nobel Prize in Medicine for his research.
Secondary structures of tRNA molecules
tRNA Structure:
- tRNA molecules are generally 75-87 nucleotides long.
- They form clover-leaf shaped structures through base pairing in the acceptor stem, D-stem, TΨC stem, and anticodon stem.
- Modified tRNA nucleosides are found universally in living organisms.
Amino acid attachement
- tRNA molecules are covalently attached to the corresponding amino acid at one end.
- At the other end, they have a triplet sequence (anti-codon) complementary to the mRNA codon.
Physical Characteristics
- tRNA has a molecular weight of ~25,000.
- The sedimentation constant is ~4 Svedberg (S) units.
- The vertical length of tRNAs is ~60Å, and the horizontal tail is also of the same length
Cloverleaf Model
- The first tRNA model, known as the ‘cloverleaf’ model, was proposed by Holley.
- The secondary structural appearance resembles trifoliate leaves of Trifolium repens (clover).
- All tRNA molecules share similar secondary structures, featuring a ‘clover-leaf’ structure with three hairpins and an acceptor stem.
Structure Details
- The acceptor stem, where the amino acid is covalently attached, is the 3′ end of the chain and terminates in the sequence 5′-CCA-3′.
- The secondary structure folds up to form a 3-dimensional structure resembling an inverted L.
L-arm Anticodon loop
- One end of the L-arm (the 3′ end of the chain) is the acceptor stem.
- The other end is the anticodon loop, which must match the codon on the mRNA.
- The distance between the two ends of the L is ~7 nm.
Ribosome Positioning
- The corner of the L is used for correct positioning on the ribosome during protein synthesis.
Stability and Distances
Tertiary structures of tRNA molecules
- In the tertiary (3-dimensional) structures of RNA, bases sometimes make hydrogen bonds with more than one partner.
- These extra hydrogen bonds compensate for the distortion in the double-stranded helical regions when the RNA folds up and help stabilize the tertiary structure.
- The covalent attachment between the tRNA and its corresponding amino-acid is achieved by another adaptor molecule, a protein molecule called aminoacyl-tRNA synthetase.
- There are at least 20 varieties of aminoacyl-tRNA synthetase, one for each kind of amino-acid.
- The synthetases recognize the detailed shape and properties of a specific amino-acid and the detailed shape of the acceptor stem in the folded tRNA molecule
- They catalyze the covalent attachment between the amino-acid and its corresponding tRNA.
- Whether it is a secondary structure or tertiary structure, at least four distinct domains or structural motifs are viewed, and they are invariably found
in all species of tRNAs. - The domains are, from 3’ end (starts with 3’ACC) of the polynucleotide chain which contain Amino acid acceptor end, TΨCG loop, Anticodon loop, DHU loop and Variable arm.
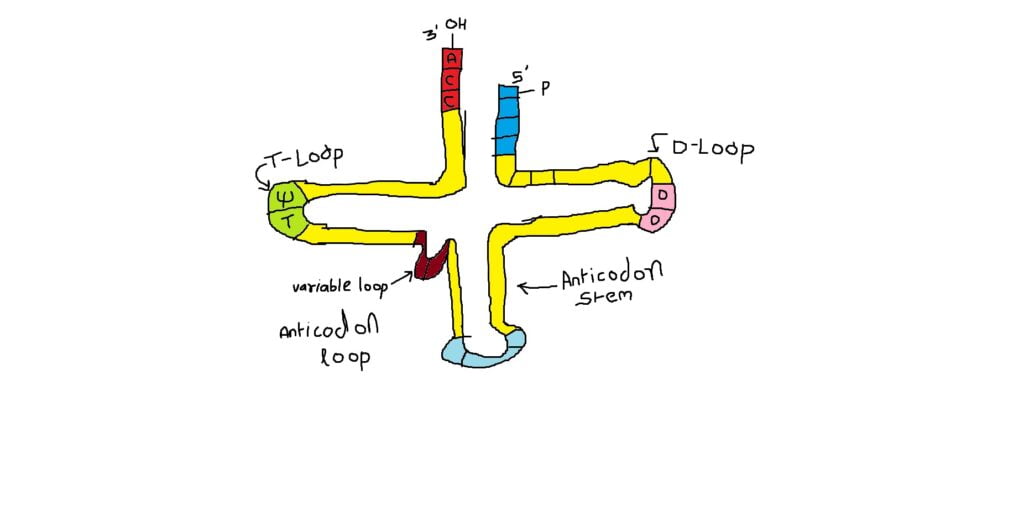
Amino Acid acceptor end
Amino acid acceptor end found at 3’, has a consensus sequence 3’ACC–, that leads to a 7 base pair stem with G: U, U: A pairings. It is to the 3’Adenine’s ribose-OH group amino acids are covalently added with carbonyl bonds. In many tRNAs 2’O is added with amino acyl group. Type I tRNAs have ACC ends as they are synthesized and Type II tRNAs don’t have when they are synthesized, but added later. The amino acid acceptor sequences may be involved in binding to a.a. tRNA synthase.
TΨCG loop
ΨCG loop is 7 bp long and consists of 3-5 bp stem. The TψCG is an invariant sequence. This region might be involved in binding of tRNA to ribosomal surface either at A or at P sites.
Anticodon loop
Anticodon loop contains seven bases with 4-5 bp long stem. Anticodons that are geometrically and stearically projected like three fingers, whose sequence differ from one species of tRNA to the other. However, more than one combination of triplet sequences as anticodons can code for the same amino acid. But very often in tRNAs, the three nucleotides, as anticodons are bracketed on either side by a modified purine (like Isopentinyl Adenine) towards 3’ side and a uracil (or pseudouridine) on 5’ side. The anticodon loop with its specific sequences may have a role in binding to amino acid tRNA synthase. Anticodons are responsible for decoding the information present in mRNAs.
DHU Loop
DHU loop ending in 5’G (in E.coli), which is paired with C in the amino acid acceptor stem. The DHU loop and its inner angle may provide specificity in tRNA and enzyme recognition.
Variable arm
Variable arm: Another additional structural feature that is found in tRNAs is an extra arm of variable length, found in between TψC stem and anticodon stem. The length varies from 2 to 21 ntds, based on the length they are classified into class I and class II tRNAs. Class I contain 3-5 ntds long arm and class-II have 13 to 21 ntds long arms.
Processing pre-tRNAs
In eukaryotes, introns are poresent in a few tRNA transcripts and must be excised. When two or more different tRNAs are contained in single primary transcript, they are separated by enzymatic cleavage mediated by endonuclease RNase P found in all organisms which removes RNA at the 5’ end of tRNAs. The 3’ end is processed by one or more nucleases, including RNase D. Many of the pre tRNAs lack 5’CAA3’ ends, in which case specific enzymes add CCA in sequence to generate the 3’ terminal A with 3’ OH group. Final type of tRNA processing is modification of some bases by methylation, deamination or reduction.
Small silencing RNAs
These are small RNA of nearly 20-30 nucleotide long which include siRNA(Small interfering RNA), miRNA(micro RNA) and piRNA (Piwi-interacting RNA).
Small interfering RNA (siRNA)
Small interfering RNA (siRNA): It is a class of double stranded RNA molecules involved in the RNA interference (RNAi) pathway. There, it interferes with the expression of specific genes controlling the stability of the mRNA. In this way the mRNA disintegrates (when necessary), avoiding its overexpression with a consequent overproduction of proteins.
Micro RNA (miRNA)
Micro RNA (miRNA) : These are endogenous regulatory RNAs which are typically 20-25 nucleotides long and are likely to regulate the expression of other genes.
Piwi-interacting RNA (piRNA)
Piwi-interacting RNA (piRNA) are a large class of small non-coding RNAs , 26-31 nucleotides in length, that interact with piwi-domain-containing proteins. piRNAs function in the germline to silence genetic elements such as retrotransposons and in RNA silencing as components of the RNA-induced silencing complex (RISC).
Small nuclear RNA (snRNA)
Small nuclear RNA (snRNA): They are RNA molecules that are found within the nucleus of eukaryotic cells.They are involved in a variety of important processes like RNA splicing (removal of introns from heterogenous nuclear RNA) and maintaining the telomeres.
Heterogeneous nuclear RNA (hnRNA)
: hnRNA is an immature single strand of mRNA. The terms hnRNA and pre−mRNA are almost identical and thus used interchangeably
Small nucleolar RNA (snoRNA):
Small nucleolar RNA (snoRNA): These are involved in chemical modifications such as methylation of rRNA and other forms of RNA in eukaryotes.
Guide RNA (gRNAs)
Guide RNA (gRNAs):These are RNa genes that function in RNA editing. RNA editing was first reported in mitochondria of Kinetoplastids,in which mRNA are edited by inserting or deleting stretches of uridylates (Us).It forms part of the editosomes that contain sequences that hybridize to matching sequences in the mRNA, to guide the mRNA modifications
tmRNA
tmRNA: It has a complex structure with tRNA like and mRNA like regions. It has currently only been found in bacteria. It recognizes ribosomes that have trouble translating or reading an mRNA and stall, leaving and unfinished protein that may be detrimental to the cell.
