From Damage to Recovery: Tracing the Pathways of DNA Repair
Author: Atharva Tilewale
Introduction
In every living organism, the DNA serves as a storehouse for genetic information, and life depends on the integrity and stability of DNA. DNA on the other hand is a chemical substance that is constantly being attacked by the environment. If this damage is not repaired, it will cause mutation and leads to illness. Skin cancer is one of the most well-known examples of the connection between environmental-induced DNA damage and disease. Skin cancer can be brought on by prolonged exposure to UV radiation from sunshine and, to a lesser extent, tanning beds. Another illustration is the harm that comes from tobacco smoking, which can induce lung cell mutations and eventually lung cancer. In addition to environmental factors, free radicals and other metabolic byproducts can cause oxidative damage to DNA. Indeed, it’s been calculated that a single cell may experience up to a million DNA alterations per day.
Apart from environmental genetic insults, mistakes can also occur in the DNA replication process itself during cell division. A key determinant of an organism’s spontaneous mutation rate is the pace at which DNA polymerase inserts incorrect nucleotides during DNA replication. Even while many of these mistakes are often identified and fixed by a “proofreading” activity of an enzyme. The frequency of mutation in DNA of human is estimated to be between 1×10-4 and 1×10-6 mutations per gamete. A rate of 1×10-6 indicates that for every million gametes, a scientist should expect to find one mutation at a certain site. In other organisms, the mutation rates are frequently far lower.
DNA Repair
Both prokaryotic and eukaryotic species contain DNA repair mechanisms, and many of the proteins involved have been conserved throughout the evolution. Indeed, cells have developed a variety of methods to identify and fix the many kinds of damage that can happen to DNA, regardless of whether replication errors or environmental factors. Since DNA is a molecule that actively participates in cell division, the management of DNA repair and the regulation of the cell cycle are intimately related.
There are two general categories of repair systems, based on the way they function.
- Direct reversal repair systems – correct damaged areas by reversing the damage
- Excision repair systems – cut out a damaged area and then repair the gap by new DNA synthesis
Direct reversal repair system includes:
- Mismatch repair by DNA polymerase proofreading
- Photoreactivation
- Alkylation repair Excision repair includes:
- Base Excision repair
- Nucleotide Excision repair
- Methyl-Directed mismatch repair
- SOS response
Direct reversal repair
1) Mismatch repair by DNA polymerase proofreading
- The frequency of base-pair substitution mutations in bacterial genes ranges from 10 -7 to 10 -11 errors per generation.
- DNA polymerase inserts incorrect nucleotides at a frequency of 10 -5 .
- DNA polymerase has 3′-to-5′ exonuclease proofreading activity in both prokaryotes and eukaryotes.
- When an incorrect nucleotide is inserted, the polymerase often detects the mismatched base pair and corrects the area by “backspacing” to remove the wrong nucleotide and then resuming synthesis in the forward direction.
2) Photoreactivation
- It is also known as light repair, it repairs UV induced pyrimidine dimers.
- Enzyme required for this mechanism is photolyase (encoded by phr gene).
- This enzyme is found in prokaryotes and simple eukaryotes (in some alga and not in humans).
- The enzyme photolyase captures the energy from the light and uses it to break the covalent bonds linking adjacent pyrimidines.
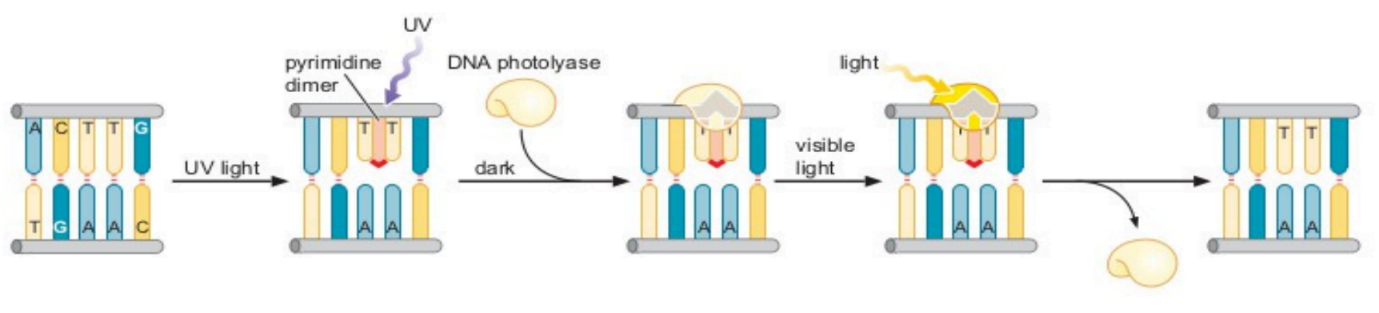
- Photolyase has 2 factors: FAD and MTHF (Methyltetrahydrofolate)
- MTHF absorbs light in the range 350-500nm. It excites and transfers e- to FAD.
- FAD is converted to FADH2. It acts on the bond between thymine (T=T ) and breaks the bond. A=T bond formation takes place immediately.
3) Alkylation repair
- Alkylating agents transfer alkyl groups (usually methyl or ethyl groups) onto the bases.
- The mutagen MMS methylates the oxygen of carbon- 6 in guanine to form O6methylguanine.
- O 6- methylguanine methyltransferase enzyme removes the methyl group by transferring it to a cysteine residue.
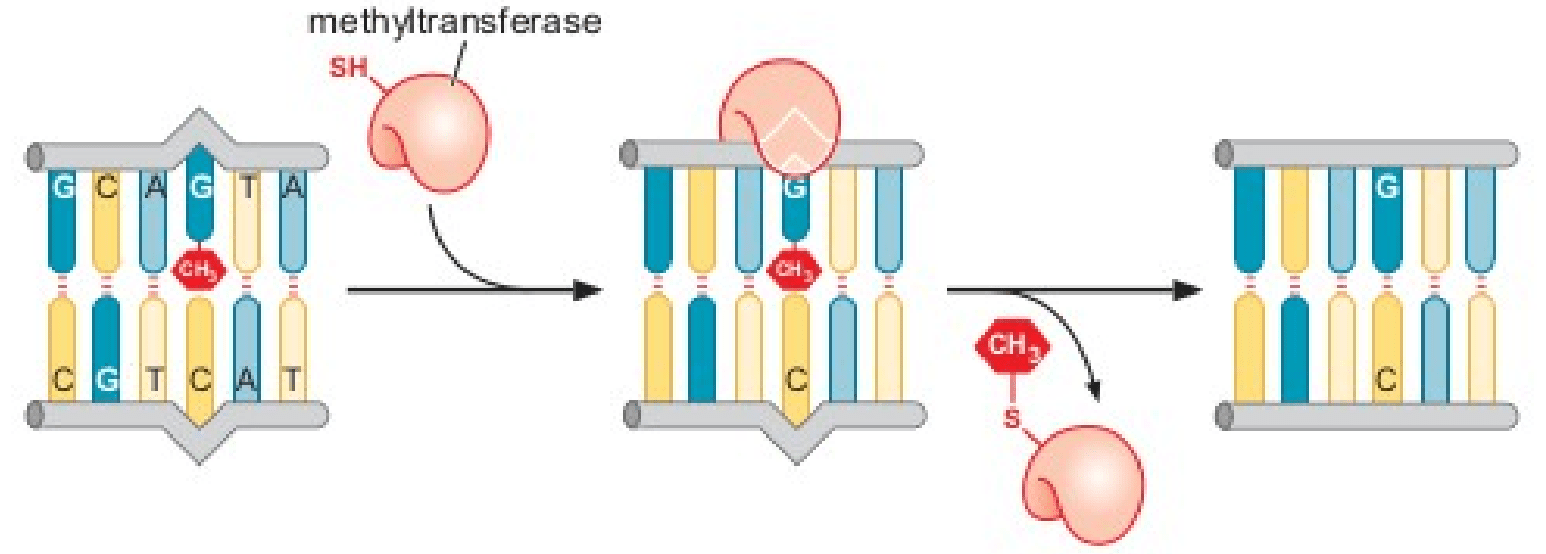
Source: James Watson, Molecular Biology of the Gene, 7th Ed.
Excision repair
1) Base excision repair
- In base excision repair, the damaged single bases are generally repaired by removing the wrong base and then inserting the correct base.
- Mutations that are caused by depurination or deamination can be corrected by the base excision repair.
- The Enzyme glycosylase helps in removing the damaged base from DNA by cleaving the glycosidic bond between base and the deoxyribose sugar. This creates the AP site. (Apurinic or Apyrimidinic site – Without purine or pyrimidine base).
- Another enzyme AP endonuclease helps in detecting the AP site and cleaves the phosphodiester bond before and after the sugar-phosphate backbone creating the gap.
- The enzyme DNA polymerase helps in filling the gap by adding correct nucleotides.
- DNA ligase later will join the nucleotide.
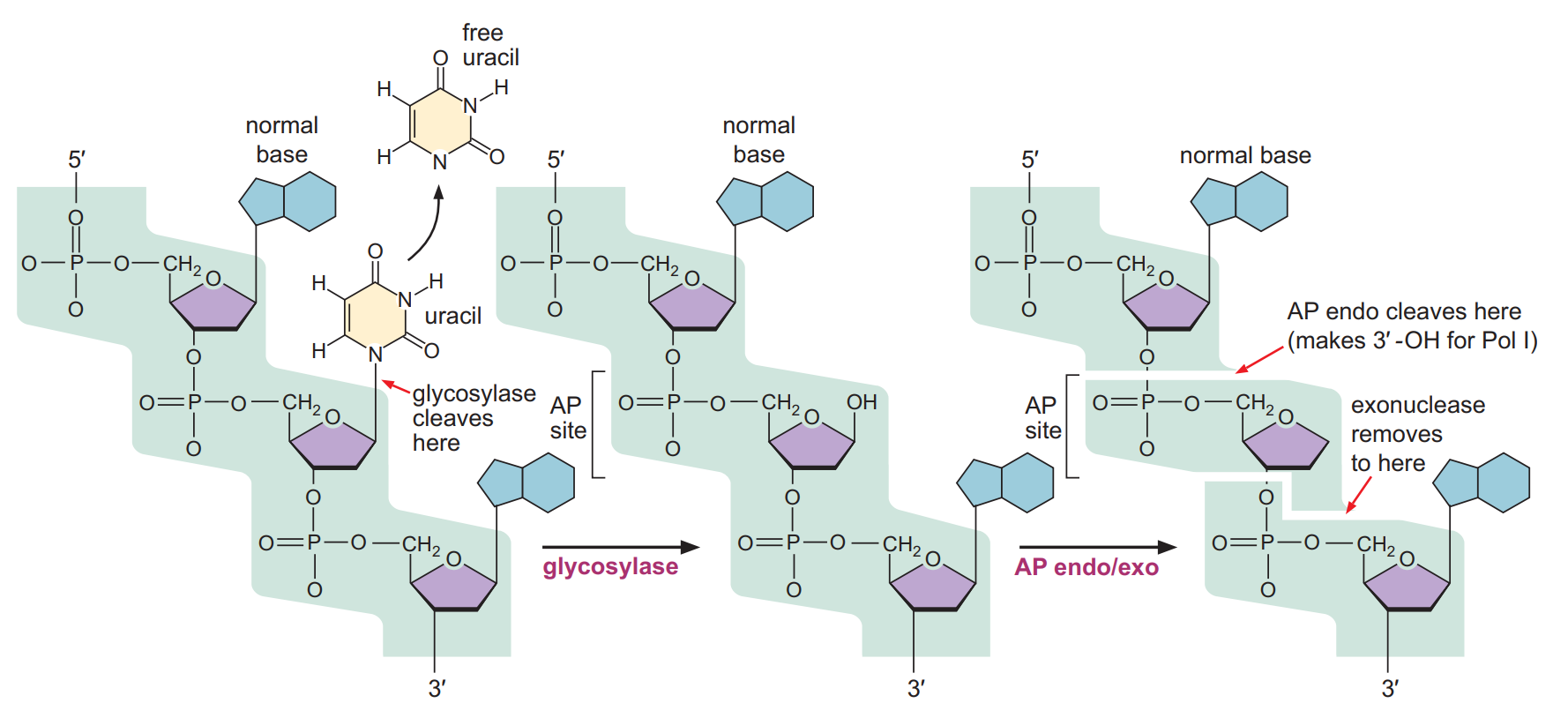
2) Nucleotide excision repair
- This system works by recognizing distortions to the shape of the double helix, such as those caused by a thymine dimer or by the presence of a bulky chemical adduct on a base.
- In this repair mechanism, the enzyme removes a short single strand segment that includes the lesion creating the gap, which is then filled by the DNA polymerase enzyme and restores the original nucleotide sequence.
- Four proteins are required: UvrA, UvrB, UvrC, and UvrD
- Two UvrA and two UvrB molecules forms a complex and scans the DNA. UvrA is detecting a distortions in the DNA.
- After detecting, UvrA leaves the complex, and the remaining dimer of UvrB melts the DNA to create a single-stranded bubble around the lesion.
- UvrB molecule dimer recruits UvrC which creates two incisions: one located 4 or 5 nucleotides 3′ to the lesion and the other 8 nucleotides 5′ to the lesion. It creates a 12 to-13 residue-long DNA strand that contains the lesion.
- UvrD which is a DNA helicase, comes and removes the nicked strand, creating a 12 to 13 nucleotide gap.
- DNA Pol I and DNA ligase comes and fills the gap.
- The principle of nucleotide excision repair in highercells is much the same as that in E. coli, but the machinery for detecting, excising, and repairing the damage is more complicated, involving 25 or more polypeptides.
3) Methyl-Directed mismatch repair
- Even after the proofreading activity by the DNA polymerase, a number of mismatched base pairs remain as it is after replication has been completed.
- Many mismatched base pairs left after DNA replication can be corrected by methyl-directed mismatch repair.
- Guanine when methylated pairs with thymine.
- This system recognizes mismatched base pairs, excises the incorrect bases, and then carries out repair synthesis.
- MutS, MutL and MutH are 3 proteins that help in repair mechanism.
- MutS binds to the mismatched region.
- MutL and MutH binds to the GATC region of daughter strand.
- Parental strand and the daughter strand is distinguished by methylated A in 5’GATC-3′ sequence.
- After replication, the parental DNA strand has a methylated A in the GATC sequence, whereas the A in the GATC of the newly replicated DNA strand is not methylated until a short time after its synthesis. Afterwards both become methylated.
- DAM methylase (Deoxyadenosine methylase) adds methyl group on the adeine in GATC sequence.
- Repair machinery acts in the buffer time i.e. between replication and methylation.
- MutS protein bound to the mismatch forms a complex with MutL and MutH, to bring the unmethylated GATC sequence close to the mismatch.
- MutH nicks the unmethylated DNA strand at the GATC site, the mismatch is removed by an exonuclease, and the gap is repaired by DNA polymerase III and ligase.
- In humans, four genes, respectively named hMSH2, hMLH1, hPMS1, and hPMS2, have been identified; hMSH2 is homologous to E. coli mutS, and the other three genes have homologies to E. coli mutL.

References
- James Watson, Molecular Biology of the Gene, 7th Ed.
- Robert Weaver, Molecular Biology, 5th Ed.
- Peter Russell, iGenetics – a Molecular Approach, 3rd Ed.
- Clancy, S. (2008) DNA damage & repair: mechanisms for maintaining DNA integrity. Nature Education 1(1):103
